DNA Fingerprinting
DNA fingerprinting of pearls was first developed by SSEF with partners at ETH Zurich and published in 2013 (Meyer et al., 2013). The method has since been further refined and the amount of required material (i.e. quasi non-destructive) has been considerably reduced. DNA fingerprinting can thus offer conclusive identification of the pearl oyster species or coral species to which a pearl or precious coral item corresponds.
Contact us for more information on this service.

DNA Fingerprinting of pearls
The Swiss Gemmological Institute SSEF has a service to support the documentation of the origin and provenance of pearls, in partnership with the Institute of Forensic Medicine at the University of Zurich. It is made possible by a substantial expansion of the organisation’s DNA fingerprinting reference database and capabilities, which now include eight oyster species that produce the vast majority of pearls found in the natural and cultured pearl trade.
The eight pearl species that can be distinguished conclusively using these DNA fingerprinting methods are:
- Pinctada radiata (Persian Gulf & Ceylon pearl oyster)
- Pinctada imbricata (Atlantic pearl oyster)
- Pinctada fucata/martensii (Akoya pearl oyster)
- Pinctada maxima (South Sea pearl oyster)
- Pinctada margaritifera (Tahitian black-lipped pearl oyster)
- Pinctada mazatlanica (Panama pearl oyster)
- Pinctada maculata (Pipi pearl oyster)
- Pteria sterna (Rainbow-lipped pearl oyster)
DNA Fingerprinting of precious corals
A breakthrough study entitled “DNA fingerprinting: an effective tool for taxonomic identification of precious corals in jewelry” in 2020 has led to a new service being offered by SSEF with IRM (University of Zürich) to aid in the traceability of precious coral jewellery.
This method uses minute amounts of DNA recovered from precious coral used in jewellery to identify their species. This is vital given that a number of precious coral species are listed on the Convention on International Trade in Endangered Species (CITES) Appendix III, and thus need to be correctly identified and declared in order to be legally traded. The ability to trace precious corals back to their species-related and geographic origins can provide greater transparency, as well as supply important scientific information for the documentation of modern and historic items.
The DNA fingerprinting technology outlined in the article represents a game-changing way of assessing the species identity of precious corals found in the trade. Importantly, the technique described here is quasi non-destructive, requires considerably less sample material than other methods, with testable DNA being recovered from as little as 2.3 milligrams (0.0115 carats) of material.
Explanation of DNA fingerprinting results for precious corals
If DNA extraction is successful and DNA fingerprinting can be carried out, SSEF can identify a coral item as one of the following:
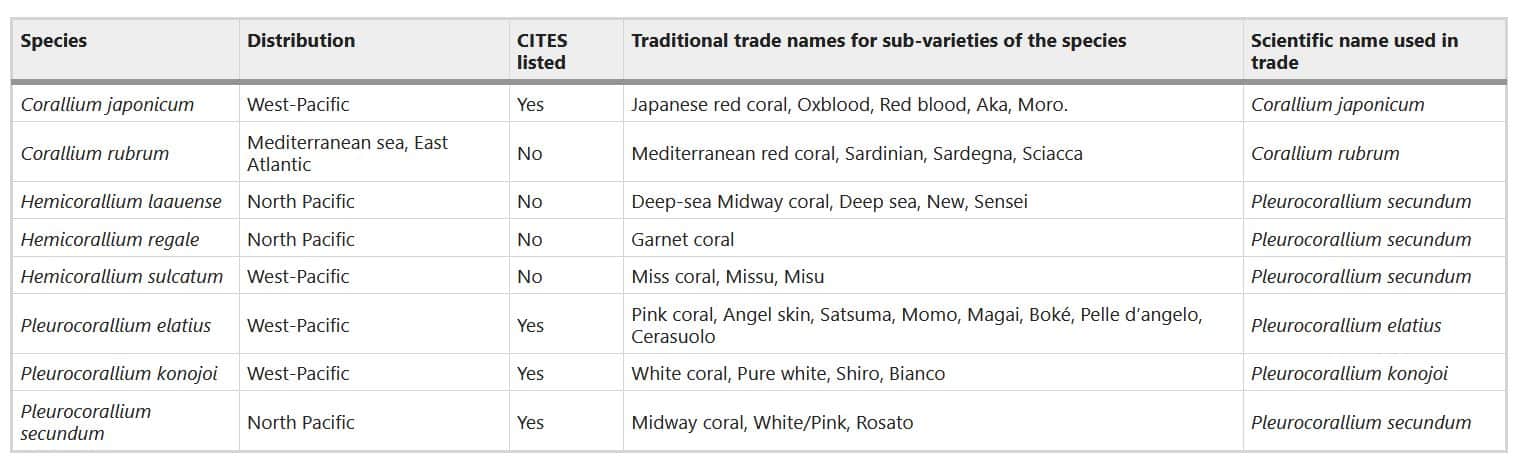
- Corallium rubrum
- Corallium japonicum complex
- Hemicorallium sp.
- Pleurocorallium secundum
- Pleurocorallium elatius complex
- Pleurocorallium sp.
Further explanation of these groups when expressed as results on a SSEF DNA fingerprinting special letter:
- Corallium rubrum: conclusive DNA species identification can be made that the sample is from Corallium rubrum (Mediterranean red coral). This species is not listed by CITES.
- Corallium japonicum complex: the following three taxa belong to this complex: Corallium japonicum, Corallium nix (white skeletal axis), Corallium tortuosum (pink skeletal axis) (sensu Tu et al. 2015). Identification of a specific species within this complex by SSEF is thus based on a combination of DNA, morphological and other analyses. Corallium japonicum (Japanese red coral) is the most widely used of these species in the jewellery trade, and includes aka, moro, oxblood varieties. Corallium japonicum is listed by CITES, the other two species within this complex are not.
- Hemicorallium sp.: This corresponds to any species within the genus Hemicorallium, and conclusive identification of the individual species is currently not possible. This genus includes deep sea Midway coral, garnet coral and Miss coral. These species are not listed by CITES.
- Pleurocorallium secundum: This species can be conclusively identified, and is known in the trade as Midway and Rosato coral. This species is listed by CITES.
- Pleurocorallium elatius complex: the following three taxa belong to the complex: Pleurocorallium elatius, Pleurocorallium konojoi and Pleurocorallium carusrubrum (red skeletal axis) (sensu Tu et al. 2015). Identification of a specific species within this complex by SSEF is thus based on a combination of DNA, morphological and other analyses. Pleurocorallium elatius is the most widely used of these species in the jewellery trade, and includes angel skin, boké, magai and momo varieties. Pleurocorallium elatius and Pleurocorallium konojoi are listed by CITES, whereas Pleurocorallium carusrubrum is not.
- Pleurocorallium sp.: any item identified as Pleurocorallium which does not belong to Pleurocorallium secundum or the Pleurocorallium elatius complex. Depending on the specific result, more exact, presumptive taxonomic identification may be possible. These species are not listed by CITES.
- no result: this means that the analysis was not successful (technical failure, likely due to too little or no DNA in the sample)
This information is based on our published research (Lendvay et al. 2020) and the phylogenetic work on precious corals by Tu et al. (2015).
DNA Fingerprinting of ivory
In 2019, SSEF became the first gem laboratory worldwide to introduce DNA fingerprinting of ivory as a standard client service. It involves a scientific method that can provide valuable information about the species of ivory being used in jewellery and ornamental objects, in order to determine whether it is CITES-listed elephant ivory or non-listed mammoth ivory.